Intro - Perceptrons
- Simplest ANN architecture
- Uses linear threshold unit (LTU) - returns weight sum of inputs, applies step function to sum, outputs result
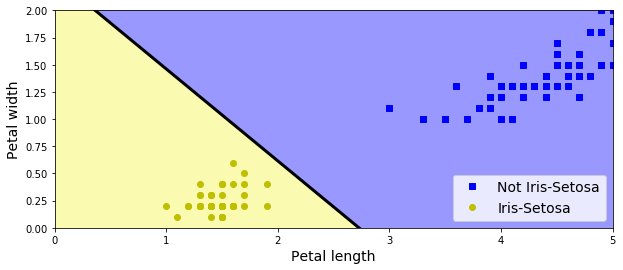
- Single LTU can be used for simple linear binary classification
- Perceptron = single layer of LTUs, each one connected to all inputs
- Percepton training based on Hebb's Rule. (basically, connection weight between two neurons goes up when they have same output.)
- Linear decision boundary, so Perceptrons not capable of learning complex patterns.
import numpy as np
from sklearn.datasets import load_iris
iris = load_iris()
X = iris.data[:, (2, 3)]
y = (iris.target == 0).astype(np.int)
from sklearn.linear_model import Perceptron
per_clf = Perceptron(random_state=42)
per_clf.fit(X, y)
y_pred = per_clf.predict([[2, 0.5]])
print(y_pred)
[1]
- Perceptron learning algo very similar go SGD.
- Perceptrons do provide class probability (like Logistic Regression classsifier). They simply make predictions based on hard threshold.
- Some limitations can be eliminated with stacked Perceptrons.
import matplotlib.pyplot as plt
a = -per_clf.coef_[0][0] / per_clf.coef_[0][1]
b = -per_clf.intercept_ / per_clf.coef_[0][1]
axes = [0, 5, 0, 2]
x0, x1 = np.meshgrid(
np.linspace(axes[0], axes[1], 500).reshape(-1, 1),
np.linspace(axes[2], axes[3], 200).reshape(-1, 1),
)
X_new = np.c_[
x0.ravel(),
x1.ravel()]
y_predict = per_clf.predict(X_new)
zz = y_predict.reshape(x0.shape)
plt.figure(figsize=(10, 4))
plt.plot(X[y==0, 0], X[y==0, 1], "bs", label="Not Iris-Setosa")
plt.plot(X[y==1, 0], X[y==1, 1], "yo", label="Iris-Setosa")
plt.plot(
[axes[0],
axes[1]],
[a * axes[0] + b,
a * axes[1] + b],
"k-", linewidth=3)
from matplotlib.colors import ListedColormap
custom_cmap = ListedColormap(['#9898ff', '#fafab0'])
plt.contourf(x0, x1, zz, cmap=custom_cmap, linewidth=5)
plt.xlabel("Petal length", fontsize=14)
plt.ylabel("Petal width", fontsize=14)
plt.legend(loc="lower right", fontsize=14)
plt.axis(axes)
plt.show()
MLPs and Backpropagation
- MLP contains one input layer, at least one hidden layer (LTU based), and one output layer (LTU based).
- Backpropagation intro'd in 1986 paper. Can be described as Gradient Descent using reverse-mode autodiff.
- For each training instance:
- Find output of each node in each consecutive layer (forward pass).
- Measure output error & how much each node in last hidden layer contributed to it.
- Measure how much each node in previous hidden layer contributed to this hidden layer.
- Repeat until input layer is reached (backward pass).
- Adjust each connection weight to reduce the error.
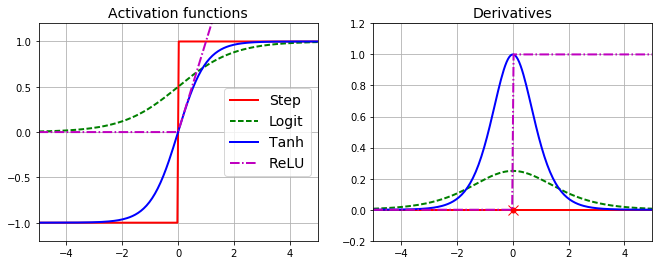
- To make algorithm work, MLP architecture changed to use logistic function delta(z) = 1/(1+exp(-z)) instead of step function.
- Backpropagation can also use hyperbolic tangent or ReLU functions if desired.
def logit(z):
return 1 / (1 + np.exp(-z))
def relu(z):
return np.maximum(0, z)
def derivative(f, z, eps=0.000001):
return (f(z + eps) - f(z - eps))/(2 * eps)
z = np.linspace(-5, 5, 200)
plt.figure(figsize=(11,4))
plt.subplot(121)
plt.plot(z, np.sign(z), "r-", linewidth=2, label="Step")
plt.plot(z, logit(z), "g--", linewidth=2, label="Logit")
plt.plot(z, np.tanh(z), "b-", linewidth=2, label="Tanh")
plt.plot(z, relu(z), "m-.", linewidth=2, label="ReLU")
plt.grid(True)
plt.legend(loc="center right", fontsize=14)
plt.title("Activation functions", fontsize=14)
plt.axis([-5, 5, -1.2, 1.2])
plt.subplot(122)
plt.plot(z, derivative(np.sign, z), "r-", linewidth=2, label="Step")
plt.plot(0, 0, "ro", markersize=5)
plt.plot(0, 0, "rx", markersize=10)
plt.plot(z, derivative(logit, z), "g--", linewidth=2, label="Logit")
plt.plot(z, derivative(np.tanh, z), "b-", linewidth=2, label="Tanh")
plt.plot(z, derivative(relu, z), "m-.", linewidth=2, label="ReLU")
plt.grid(True)
plt.title("Derivatives", fontsize=14)
plt.axis([-5, 5, -0.2, 1.2])
plt.show()
def heaviside(z):
return (z >= 0).astype(z.dtype)
def sigmoid(z):
return 1/(1+np.exp(-z))
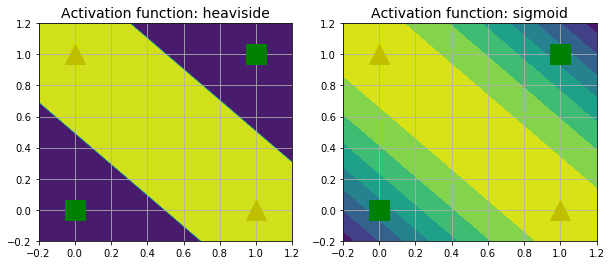
def mlp_xor(x1, x2, activation=heaviside):
return activation(
-activation(x1 + x2 - 1.5) + activation(x1 + x2 - 0.5) - 0.5)
x1s = np.linspace(-0.2, 1.2, 100)
x2s = np.linspace(-0.2, 1.2, 100)
x1, x2 = np.meshgrid(x1s, x2s)
z1 = mlp_xor(x1, x2, activation=heaviside)
z2 = mlp_xor(x1, x2, activation=sigmoid)
plt.figure(figsize=(10,4))
plt.subplot(121)
plt.contourf(x1, x2, z1)
plt.plot([0, 1], [0, 1], "gs", markersize=20)
plt.plot([0, 1], [1, 0], "y^", markersize=20)
plt.title("Activation function: heaviside", fontsize=14)
plt.grid(True)
plt.subplot(122)
plt.contourf(x1, x2, z2)
plt.plot([0, 1], [0, 1], "gs", markersize=20)
plt.plot([0, 1], [1, 0], "y^", markersize=20)
plt.title("Activation function: sigmoid", fontsize=14)
plt.grid(True)
plt.show()
MLP Training
- MLP often used for classification - each output corresponding to distinct binary class (ex: urgent/not-urgent, spam/not-spam, ...)
- If exclusive classes, output layer often uses shared softmax function.
DNN Training with "plain" TF
- Use mini-batch gradient descent on MNIST dataset
- Specify #inputs, #outputs, #hidden neurons in each layer
import tensorflow as tf
tf.reset_default_graph()
n_inputs = 28*28
n_hidden1 = 300
n_hidden2 = 100
n_outputs = 10
learning_rate = 0.01
X = tf.placeholder(tf.float32, shape=(None, n_inputs), name="X")
y = tf.placeholder(tf.int64, shape=(None), name="y")
'''
No need to define your own. TF shortcuts:
fully_connected()
'''
def neuron_layer(X, n_neurons, name, activation=None):
with tf.name_scope(name):
n_inputs = int(X.get_shape()[1])
stddev = 1 / np.sqrt(n_inputs)
init = tf.truncated_normal((n_inputs, n_neurons), stddev=stddev)
W = tf.Variable(init, name="weights")
b = tf.Variable(tf.zeros([n_neurons]), name="biases")
Z = tf.matmul(X, W) + b
if activation=="relu":
return tf.nn.relu(Z)
else:
return Z
with tf.name_scope("dnn"):
hidden1 = neuron_layer(X, n_hidden1, "hidden1", activation="relu")
hidden2 = neuron_layer(hidden1, n_hidden2, "hidden2", activation="relu")
logits = neuron_layer(hidden2, n_outputs, "output")
with tf.name_scope("loss"):
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(
labels=y,
logits=logits)
loss = tf.reduce_mean(
xentropy,
name="loss")
with tf.name_scope("train"):
optimizer = tf.train.GradientDescentOptimizer(learning_rate)
training_op = optimizer.minimize(loss)
with tf.name_scope("eval"):
correct = tf.nn.in_top_k(
logits, y, 1)
accuracy = tf.reduce_mean(
tf.cast(correct, tf.float32))
init = tf.global_variables_initializer()
saver = tf.train.Saver()
Execution Phase
- Load MNIST using TF helpers (fetch, auto-scale, shuffle, provide minibatch function)
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("/tmp/data/")
n_epochs = 20
batch_size = 50
Extracting /tmp/data/train-images-idx3-ubyte.gz
Extracting /tmp/data/train-labels-idx1-ubyte.gz
Extracting /tmp/data/t10k-images-idx3-ubyte.gz
Extracting /tmp/data/t10k-labels-idx1-ubyte.gz
with tf.Session() as sess:
init.run()
for epoch in range(n_epochs):
for iteration in range(mnist.train.num_examples // batch_size):
X_batch, y_batch = mnist.train.next_batch(batch_size)
sess.run(
training_op,
feed_dict={X: X_batch, y: y_batch})
acc_train = accuracy.eval(
feed_dict={X: X_batch, y: y_batch})
acc_test = accuracy.eval(
feed_dict={X: mnist.test.images,
y: mnist.test.labels})
print(epoch, "Train accuracy:", acc_train, "Test accuracy:", acc_test)
save_path = saver.save(sess, "./my_model_final.ckpt")
0 Train accuracy: 0.94 Test accuracy: 0.8753
1 Train accuracy: 0.9 Test accuracy: 0.9087
2 Train accuracy: 0.94 Test accuracy: 0.9212
3 Train accuracy: 0.88 Test accuracy: 0.9239
4 Train accuracy: 0.88 Test accuracy: 0.9335
5 Train accuracy: 0.96 Test accuracy: 0.9365
6 Train accuracy: 0.92 Test accuracy: 0.9415
7 Train accuracy: 0.94 Test accuracy: 0.9449
8 Train accuracy: 0.96 Test accuracy: 0.9459
9 Train accuracy: 0.94 Test accuracy: 0.9497
10 Train accuracy: 1.0 Test accuracy: 0.9539
11 Train accuracy: 0.94 Test accuracy: 0.9563
12 Train accuracy: 0.98 Test accuracy: 0.958
13 Train accuracy: 0.98 Test accuracy: 0.9584
14 Train accuracy: 0.94 Test accuracy: 0.9614
15 Train accuracy: 1.0 Test accuracy: 0.9622
16 Train accuracy: 0.96 Test accuracy: 0.9639
17 Train accuracy: 0.92 Test accuracy: 0.9635
18 Train accuracy: 0.98 Test accuracy: 0.9654
19 Train accuracy: 0.92 Test accuracy: 0.9667
Using in production
- Now trained - you can use the NN to predict.
with tf.Session() as sess:
saver.restore(sess, save_path)
X_new_scaled = mnist.test.images[:20]
Z = logits.eval(feed_dict={X: X_new_scaled})
print(np.argmax(Z, axis=1))
print(mnist.test.labels[:20])
[7 2 1 0 4 1 4 9 6 9 0 6 9 0 1 5 9 7 3 4]
[7 2 1 0 4 1 4 9 5 9 0 6 9 0 1 5 9 7 3 4]
Parameter Tuning
- Way too many parameters - Grid Search approach not time-effective.
- 1st option: randomized search.
- 2nd option: Oscar
- Start with common defaults to restrict search space.
Number of hidden layers
- Deep nets have much better parameter efficiency than shallow ones. (They can model complex functions with much fewer neurons.)
- Largely due to hierarchical nature of most data modeling probs
Number of neurons per hidden layer
- Determined by input dimensions. Ex: MNIST requires 28x28 inputs, 10 outputs
- Try increasing # of layers before # neurons/layer.
- Simple trick: pick model w/ excessive layers & neurons, use early stopping, regularization, dropout, etc. to prevent overfit.
Activation functions
- Defaults:
- use ReLU in hidden layers. Faster & helps avoid GD getting stuck on local plateaus.
- use Softmax in output layer (for classification; none needed for regression.)


